Startling Evidence suggests BioNTech/Pfizer Falsified Key Data & Further Scandals (part 2)

Originally published on Trial Site News
Part 1 of my investigative report published in Trial Site News, focused on the unusual-looking Pfizer-BioNTech “Western Blots” – the scandal known as Blotgate. Western Blots were performed by BioNTech to prove the fidelity of their product (to regulators) that only the expected spike protein was expressed in vitro by modified vaccine mRNA and that it was consistent between different batches.
The problem with the fact that these don’t look like authentic/conventional Western blots, but rather computer-generated versions (automated Western blots, although not labeled as such in any of the reports), is a distraction from the real scandal at hand: the evidence, which shows, on its face, that BioNTech/Pfizer “copied and pasted” these strips, in other words, falsified their key data. These apparently fabricated Western strips (shown below) were exposed in Pfizer’s response to questions from the US Food and Drug Administration around November 2020, in the run-up to emergency use approval.
Only when these bands were quantified using image software analysis (thanks to an anonymous expert) did the “copy and paste” work, performed on 4 separate batches of their product, transfected at 6 different concentrations, become strikingly visible.
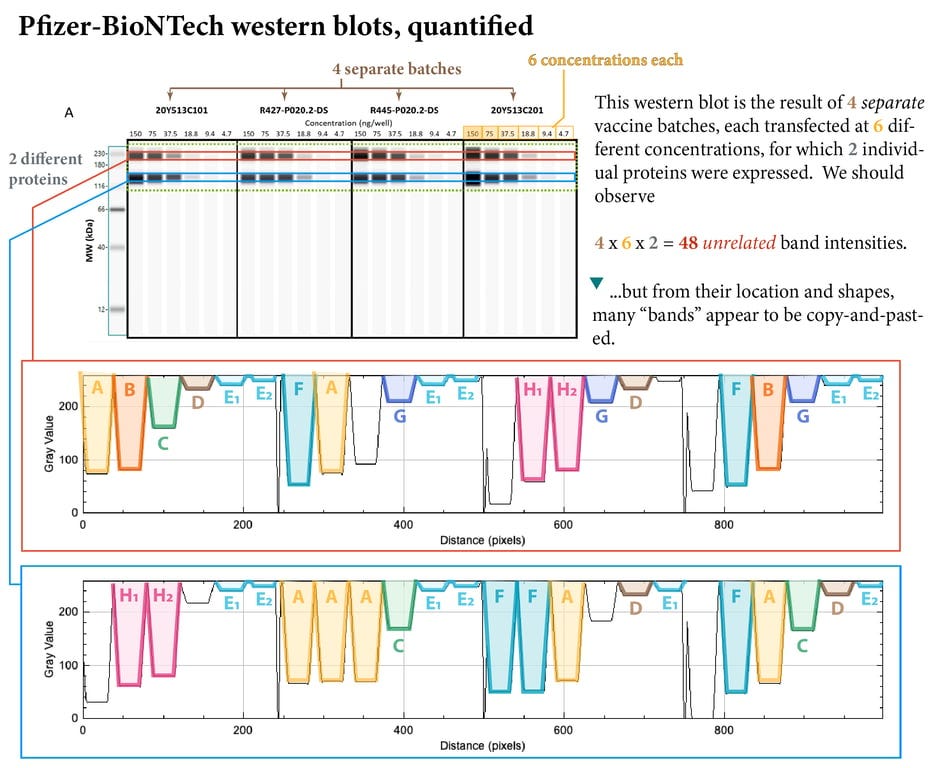
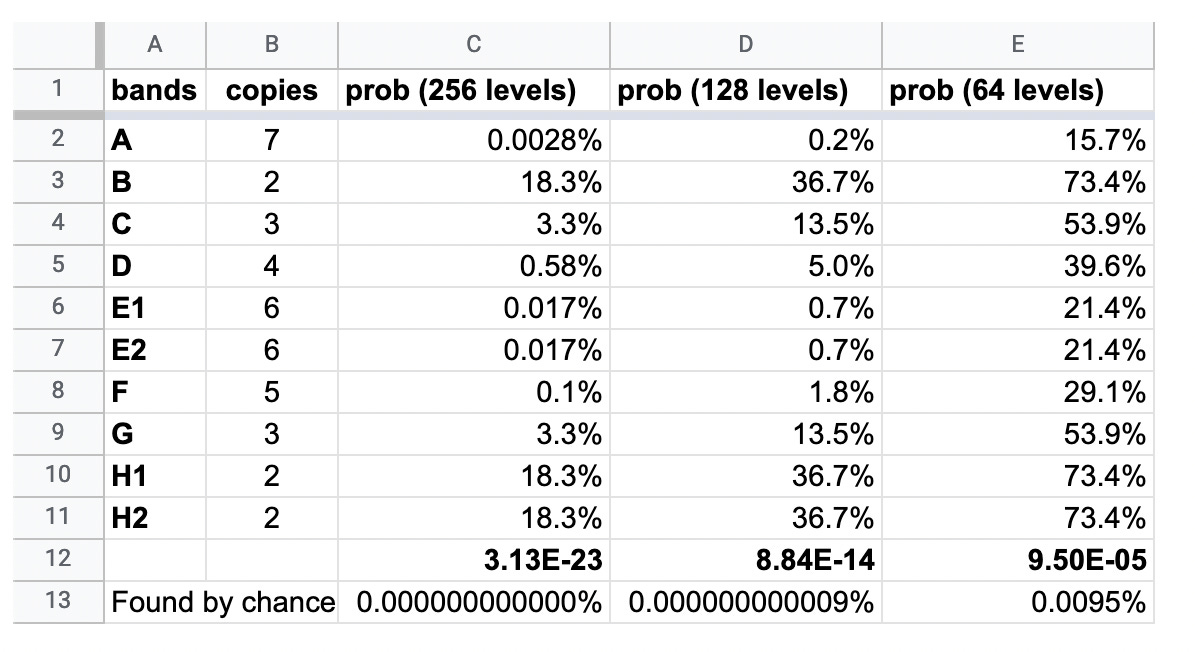
The chances of the exact same bands occurring naturally in four separate batches of vaccine, transfected at 6 different concentrations, can be seen in the table below.
On the heels of Part 1 of my investigative report, Epoch Times conducted its own investigation into Blotgate; Pfizer-BioNTech vaccine mRNA quality issues and referenced the Trial Site News investigation into the European Medicines Agency (EMA) email and document leaks.
‘Hiding the dirty tapes’ – a further look at Blotgate
On January 13, shortly after the Blotgate scandal broke, the paper sponsored by Pfizer and BioNTech, Patel et al. entitled “Characterization of BNT162b2 mRNA to Evaluate Risk of Off-Target Antigen Translation” was published on in the Journal of Pharmaceutical Sciences. The paper has many anomalies and it is worth noting that computational and molecular biologist, Dr. Jessica Rose, has written a comprehensive critical analysis of this study.
First, it is worth noting the authors’ competing interests, which can be seen below.

In speaking with Dr. Jessica Rose, who has extensively performed conventional Western Blots, she explained, “In my expert opinion, [a traditional Western] is one of the bench tests that is a procedure, like biologics safety testing (which is supposed to take 10 years). A Western blot requires a sequence of specific steps, these steps cannot be rushed and cannot overlap… experiments performed using Robo Jess [automated Western blot machine] and blots submitted by Pfizer must be reproducible by human hands, reproducibility is a requirement, especially when the list of conflicts of interest is so long on the part of the submitting authors [Patel et al.].””
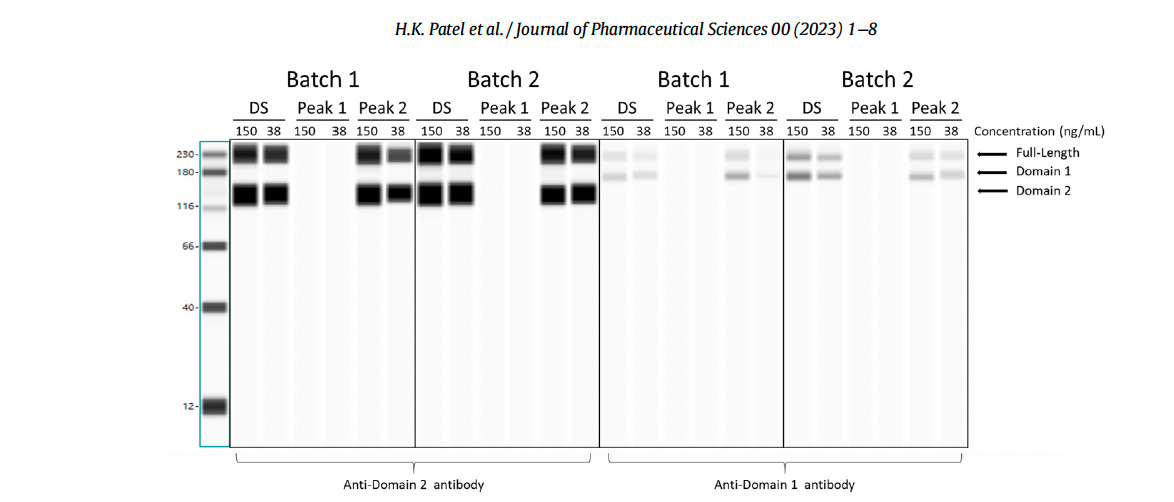
The image below, taken from the Patel et al. paper shows their “Western Blot” Notice the very thick (homogeneous) black regular bands, without any smears.
The same “Western Blot” (seen below), only a much cruder, copy-scanned version can be found in the redacted European Medicines Agency CHMP (Committee for Medicinal Products for Human Use) report from August 2021, about 18 months before the Patel et al. paper.
Since the authors Patel et al. refer to ProteinSimple technology in their study (excerpted below), a comparison can be made with an automated Western blot sample using the same company’s software.
‘Cell lysates were analyzed with specific antibodies to detect SARS-CoV-2 spike proteins using ProteinSimple technology. 12-230 kDa Wes Separation Module and 25 capillary cartridges were used. Mouse SARS-CoV-2 Spike S1 subunit antibodies (R&D systems, catalog no. MAB105403) and mouse SARS-CoV-2 Spike S2 subunit antibodies (R&D systems, catalog no. MAB10557) were used. mRNA sample results are reported as images of the ProteinSimple Wes
Source: Patel et al
The image below represents an automated Western using ProteinSimple technology.
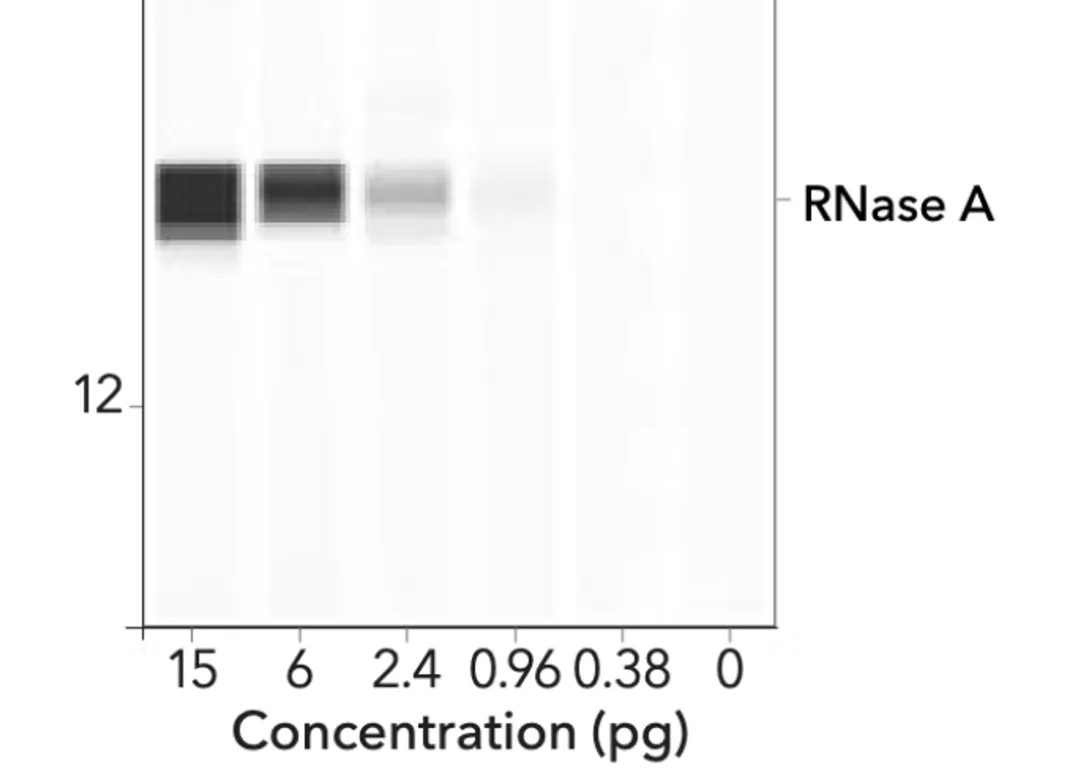
What is striking is that a gradient (smeared effect) can be seen in each of the varied bands. These look quite different from the thick, homogeneous black bands of BioNTech/Pfizer.
To get more information, we interviewed Kevin McKernan, the Human Genome Project’s R&D leader and genomics scientist, who explained that “ the BioNTech/Pfizer data is unusable” and that his reasoning for the unusual appearance of the Western Blot strips (noted in the EMA report and the Patel paper) is because “they [BioNTech] have greatly increased the gain and most people do this to hide the ‘dirty strips’ There are probably 5 strips there [in one thick strip that looks unique]” I refer to this informative interview later in this report.
This begs the question: did BioNTech/Pfizer perform an intentional cover-up of their “Western blot” results by submitting manipulated versions (of automated Westerns) to regulators? Perhaps it was copy and paste work for the FDA and manipulation of saturation levels for the EMA? And more importantly: how did the regulators accept these “Western blots” as the primary evidence to demonstrate the fidelity and consistency of BioNTech and Pfizer’s product?
We have contacted the EMA for a response to the concerns raised in this investigation. Their press officer responded with the following statement: ‘These figures [Western blot images] were extracted from the dossier submitted and inserted into the assessment report, which resulted in a loss of image quality. In addition, the redaction software used by the EMA to prepare documents for publication following an ATD request has an impact on the resolution of the documents.
In terms of method validation, the Western Blot analytical methods used in the characterisation studies were previously evaluated during the initial conditional marketing authorisation application as part of the ‘development history and comparability assessment’ and were considered appropriate for this purpose
FDA, Pfizer and BioNTech did not respond for comment.
Quality parameters established months after marketing authorisation was granted
The reason for the marketing authorisation holder (BioNTech) to conduct an additional series of tests was that the conditional marketing authorisation (CMA) was granted on the basis of the marketing authorisation holder fulfilling specific obligations (such as additional data to further characterise the truncated and modified mRNA species) imposed by the EMA. By the time the MA was granted, several CMC (chemistry, manufacturing and control) issues had been raised as major concerns by the EMA, in particular the decrease in mRNA integrity (presence of truncated/fragmented mRNA – lacking a critical attribute, a 5′ end and/or poly(A) tail) of commercial batches compared to those used in clinical trials. the “solution” to this major problem was to reduce the acceptance criteria for fragmented/truncated mRNA specimens to 50%, which the regulators simply dropped.
Canadian pharmacist, Maria Gutschi, PharmD, who has over 30 years of hospital, community and government experience, independently reviewed the Pfizer/BioNTech vaccine quality issues identified by the European Medicines Agency in an informative video presentation. Speaking to Gutschi, she raised other important concerns: ‘It was not until May 2021 [5 months after the authorisation was granted] that the OCABR established the quality parameters and, more importantly, the standardisation of the tests used. One issue we found was that, at launch, many of the tests used to determine quality and identity were ‘in-house’ tests by BioNTech. Well, a regulatory body cannot accept that. They need to be validated so that they are consistent, reproducible and reliable‘
OCABR stands for Official Control Authority for Batch Release, which sets guidelines for authorised human vaccines in the EU. Note the May 2021 date highlighted in the OCABR document, seen below.

The fact that the OCABR has set the first quality parameters for a human vaccine, an astonishing 5 months after the CMA was granted- is unprecedented. Secondly, the fact that these tests (assays) had to be validated is noteworthy.
Questionable “in-house” tests
It is worth noting that Gutschi’s concern about BioNTech’s “in-house” testing was also shared by an EMA reviewer in the leaked November 2020 Continuing Reporter Review Report shortly before the CMA was granted.
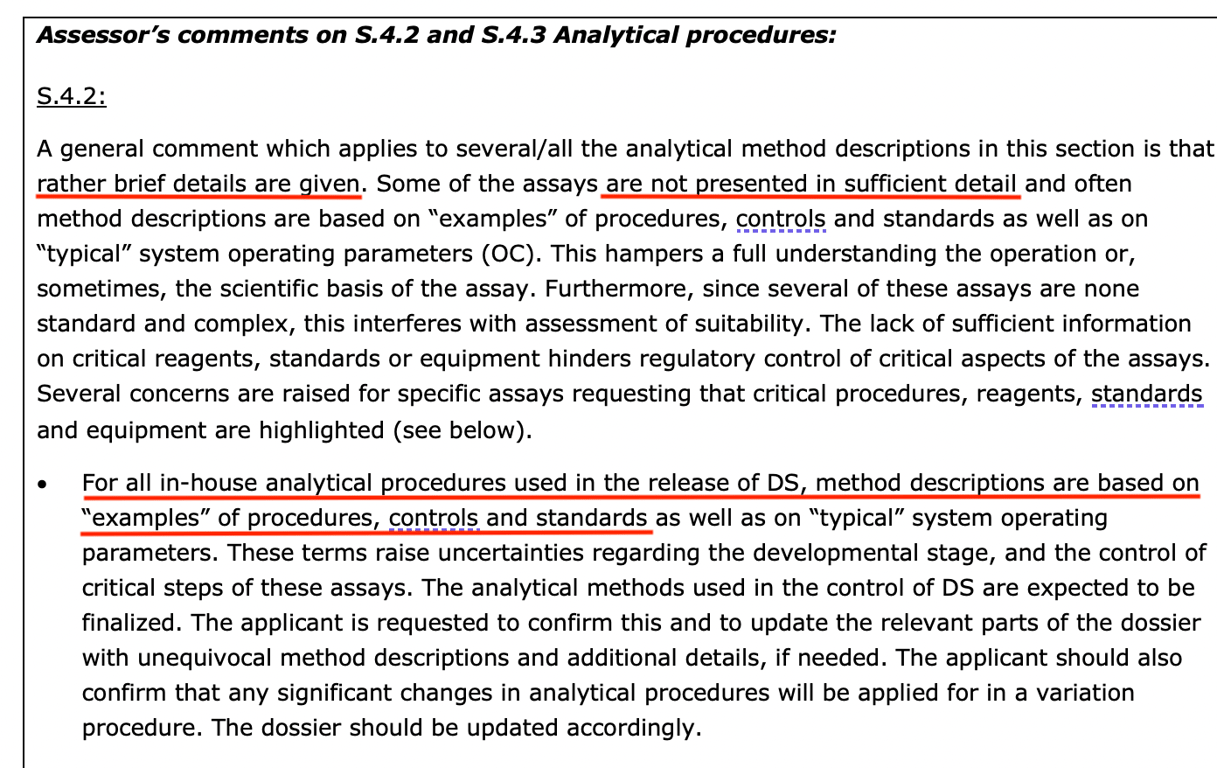
Referring back to FDA’s 2016 guidance document on “Data Integrity and CGMP Compliance,” it states, ” Data Integrity and CGMP Compliance: “In recent years, FDA has increasingly observed CGMP violations involving data integrity during CGMP inspections. This is concerning because ensuring data integrity is an important component of industry’s responsibility to ensure the safety, efficacy and quality of drugs and FDA’s ability to protect public health”
The FDA’s consideration of data integrity breaches as “concerning” seems to be disappearing – by readily accepting BioNTech’s seemingly “copy and paste” data. Perhaps for them it was just a box-ticking exercise to make it look like they were doing their due diligence to “protect public health”
“Unexpected” bands appearing in BioNTech’s genuine Western blots
Among all the computer-generated “Western blots”, BioNTech actually submitted two authentic Westerns (the second will be discussed later), which proved they knew how to make them. In the leaked EMA reporter’s “Rolling Review” evaluation report, the following criticisms of BioNTech’s 20-0211 study were made“with respect to Western blot results”, noted below.
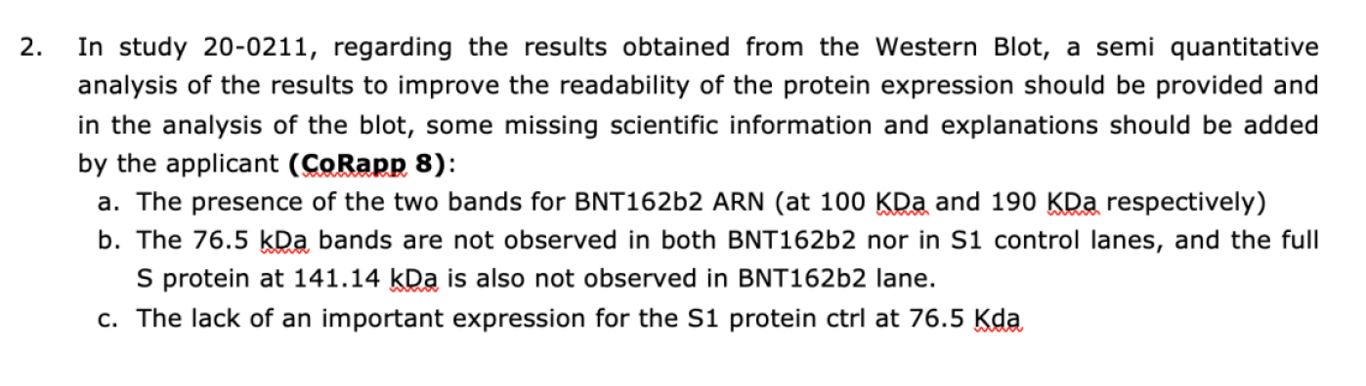
Last February, study 20-0211 (the study specified above) conducted by BioNTech was published as part of the court-ordered Pfizer data dump. The highly revealing authentic Western Blot shown in that study can be seen below.

From the EMA’s November 2020 assessment report, we know that the agency flagged the unexpected molecular weights (measured in kDa) of the two protein bands shown in the Western Blot, 190kDa and 100kDa respectively, prompting it to ask for an explanation from BioNTech. Now, the full spike protein has a molecular weight of 141 kDa, and S1 (spike protein subunit) has a molecular weight of 76.5 kDa, which was used as a control. Notice from the figure above that there are no protein bands at any of these expected molecular weights in the BNT 162B2 band and that the expected 76.5 kDa molecular weight of the S1 protein was not observed in the S1 control band. These anomalies stood out to the regulator, but somehow the vaccine developer, BioNTech, who would surely have known the expected molecular weight of the full spike protein since its mRNA product was supposed to encode it, didn’t even recognize them.
To exacerbate their complete failure to address these issues, BioNTech wrote a wildly inaccurate description of their Western Blot – in fact, the exact opposite of what was shown in the picture: “BNT162b2b2 has an expected size of 141.14 kDa” and “the S1 subunit protein of SARS-CoV-2 (76.5kDa) was used as a positive control“
This suggests that it is likely that the S1S2 spike protein may not have been expressed by the vaccine-modified mRNA, or if it was, perhaps other aberrant proteins were also expressed that originate from fragmented/truncated RNA molecules in the drug substance.
Lack of genome sequencing and N1 methylpseudourin issues
When asked about the problems with the quality of the Pfizer-BioNTech vaccine, genomics researcher Kevin McKernan gave the following response:
“We don’t have any DNA sequencing on these batches. This is absolute madness! Coming from the human genome project, we release a sequence every 24 hours to make sure the world has access to the latest data coming from the human genome project. Fast forward to today and you can’t find any sequencing of genome batches“
It’s amazing that regulators have relied on these manipulated-looking Westerns blot tests from BioNTech, instead of requiring genome sequencing of batches/lots to prove their consistency and fidelity.
McKernan went on to address the problems caused by altered RNA synthesized using an altered base:“We have an mRNA product in which every uridine has been replaced with N1 methylpseudourine, which the body has never seen before. They [BioNTech and Pfizer] chose stop codons that are the most notorious for creating errors. They [were aware of the problem but didn’t fix it properly. This means that when ribosomes go to read the template, they are very confused because they have never seen it before.”
McKernan co-authored a paper with Dr. Peter McCullough and Anthony Kyriakopoulos entitled “Differences in Vaccine and SARS-CoV-2 Replication Derived mRNA: Implications for Cell Biology and Future Disease” The authors concluded that “changes in synonymous codons embedded in mRNA vaccines may alter the expected conformation of the encoded protein, as the speed and efficiency of translation may lead to a different protein fold… Codon optimization strategies for mRNA vaccine development may lead to immune dysregulation, affect epi-transcriptomic regulation and lead to disease progression“
Modified uridine (N1 methylpseudorin) has been incorporated into mRNA to circumvent the innate immune response and promote protein translation. However, in the EMA’s November 2020 Rolling Review report (page 61), a potential safety risk with modified RNA (modRNA) was raised, see below.
‘MODRNA contains a 1-methyl-pseudouridine substitution with uridine. This substitution diminishes the recognition of the vaccine RNA by innate immune sensors. However, no further discussion of the risk of modRNA-induced autoimmune responses was provided. The applicant is invited to further discuss the possibility that the mRNA vaccine may trigger potential autoimmune responses and how they intend to assess the possible occurrence of such responses
It is not known whether BioNTech has ever assessed the potential safety risk of autoimmune response caused by either modified uridine or translated proteins (other than spike protein), as as of August 2021, with the July 2021 deadline passed, this has not yet been done. (See snapshot below, page 13 of EMA’s August 2021 CHMP report)

What is disturbing are the findings of the largest study of its kind from King Fahad University Hospital in Khobar, Saudi Arabia, linking mRNA vaccines to the triggering of autoimmune diseases, which TrialSite just recently reported. “Molecular mimicry,” stated as a concern in the EMA report above, is the same hypothesis advanced in the study as the mechanism associated with mRNA vaccines causing an autoimmune process.
Potential problems arising from the manufacturing process
The figure below shows the simplistic view used to promote how mRNA vaccines work.
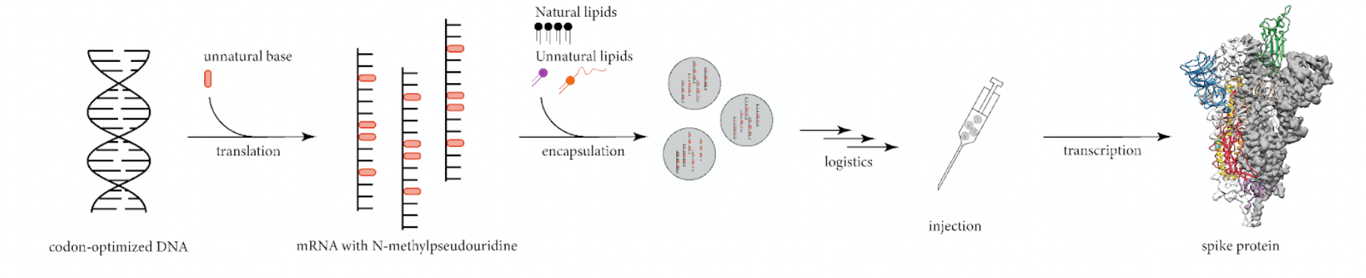
When in fact the process is highly variable, which in turn generates highly variable protein products, see figure below.

Each step of the manufacturing process can introduce unknown impurities and unknown errors, from the “unfaithful” translation of codon-optimised DNA, which can generate (non-intact) mRNA fragments, to the “unfaithful” transcription of the mRNA mixture in the human body, which can generate unexpected protein products.
McKernan went on to further explain the error rate and its implications: ‘My estimate is that there is one error per molecule of vaccine – given that there are 14 trillion molecules in each injection – we don’t know what that means immunologically. That’s why there has to be batch-by-batch sequencing before injecting a product known to produce new proteins inside humans‘
The full-length modified mRNA of the Pfizer-BioNTech vaccine encoding the spike protein is ∼4300 nucleotides long, nt. Anything less is considered a fragmented mRNA species, which EMA has classified as a “product-related impurity”. This leads to another scandal, the one known as Humpgate.
The Humpgate scandal
Humpgate can be seen as the precursor to Blotgate. While Blotgate focused mainly on the falsely expressed protein bands of vaccine mRNA observed in BioNTech Westerns, Humpgate refers to the truncated mRNA species reported by the EMA and other regulators. The name comes from the “humps” seen in the image in the social media post below.
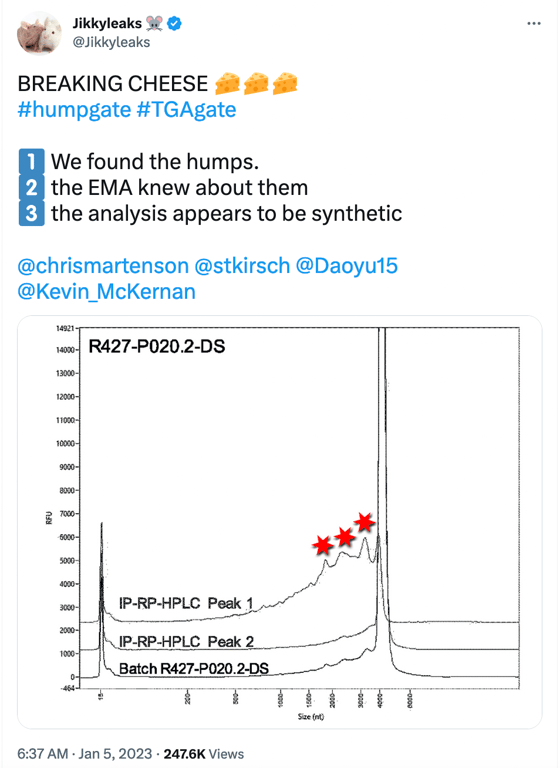
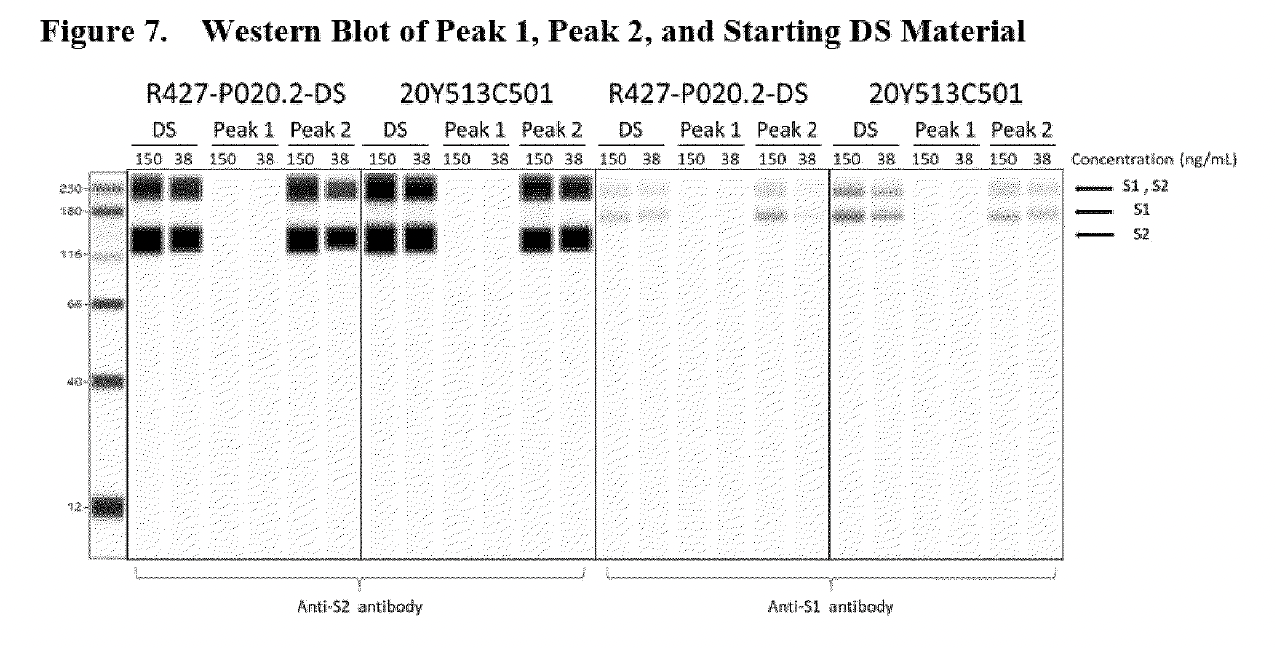
The 3 “humps” indicated by the red stars represent truncated/fragmented RNA species having a shortened length (nt) <4000nt. The image above is a Fragment Analyser electropherogram (which is the top image in Figure 2 shown below) taken from page 15 of the EMA report written in summer 2021, which BioNTech undertook to meet SO1 (specific obligation 1) set by the EMA, which requested additional data to better characterise truncated and modified mRNA species.

Fractionated samples were taken from Trial 1(R427-P020.2-DS, clinical trial batch, top image) and Trial 2(20Y513C501, PPQ batch, bottom image) using RP-HPLC with ion pairing to further characterize intact (full-length) and fragmented mRNA species. Both batches (Trial 1 and 2) exhibit small bumps leading to a large peak at a size of approximately 4300 nt, which represents the full-length mRNA species. The graphic line labeled as Peak 1 shows the “purified” reanalyzed material composed of fragmentary mRNA species only (these are the small bumps observed on the left side of the large peak of approximately 4300 nt in the batch sample), and Peak 2 is the “purified” reanalyzed material composed of intact mRNA species observed at the “large peak”
The same electropherogram can be found in the paper published in January 2023, Patel et al. (see image below, published 18 months after the EMA report). However, authors Patel et al. cite their research as current, when it is a reworked version of Pfizer/BioNTech’s own response to FDA and EMA questions from 18 months ago. The fact that the authors presented their study as “independent” and “current” when the ProteinSimple Wes technology used in their research has been discontinued as of July 30, 2021, is disingenuous to say the least.
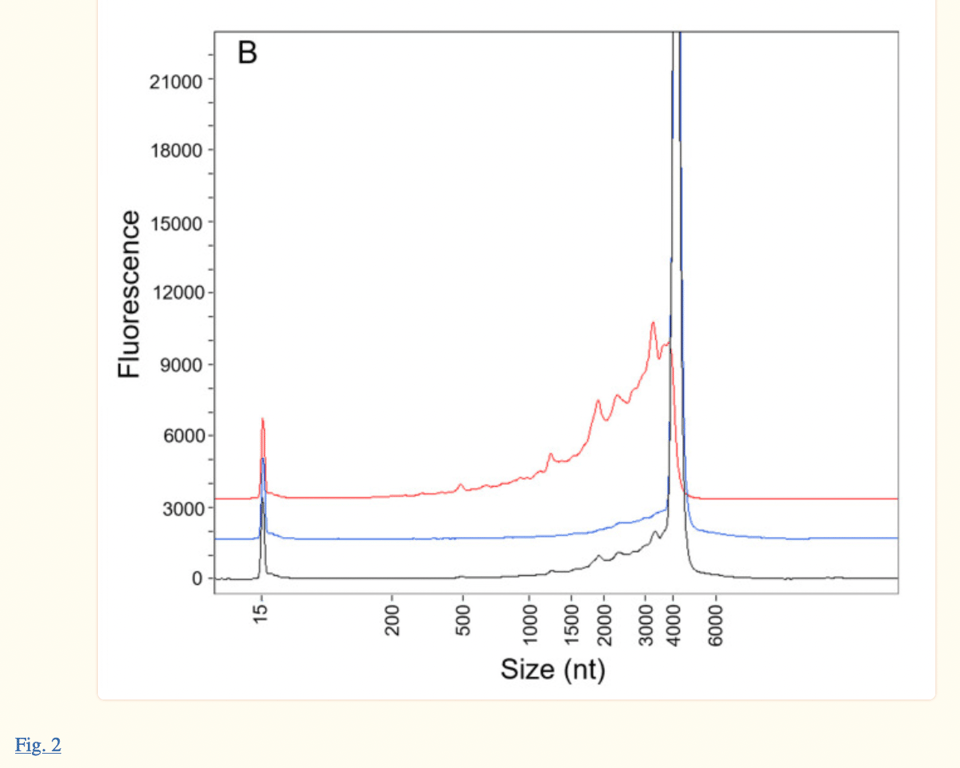
Both the Patel et al. paper and the EMA paper take issue with the humps observed on the left side of the main peak at about 4300 nt. The EMA report states “(Figure 2) demonstrates that Peak 1 consists almost entirely of fragmented species, consistent with data previously provided in the Assessment of Response to CHMP Q01-Quality 11-Dec-2020”
The problem with the BioNTech hypothesis
The way BioNTech assured EMA that these truncated RNA species would not be able to support protein translation, so that the truncated species seen as “humps” in electropherograms would not be considered a problem, was by performing new Western Blot assays to prove that RNA “transcripts require both 5′-cap and poly(A) to support protein translation…”

Here’s what a full-length BNT162b2 RNA looks like. A 5′-cap is added to the beginning of the RNA transcript, and a 3′-poly(A) tail is added to the end of the transcript. Ribosomes read the transcript from the 5′ to the 3′ direction.

The figures below (6 and 5) are taken from the EMA CHMP report of August 2021.
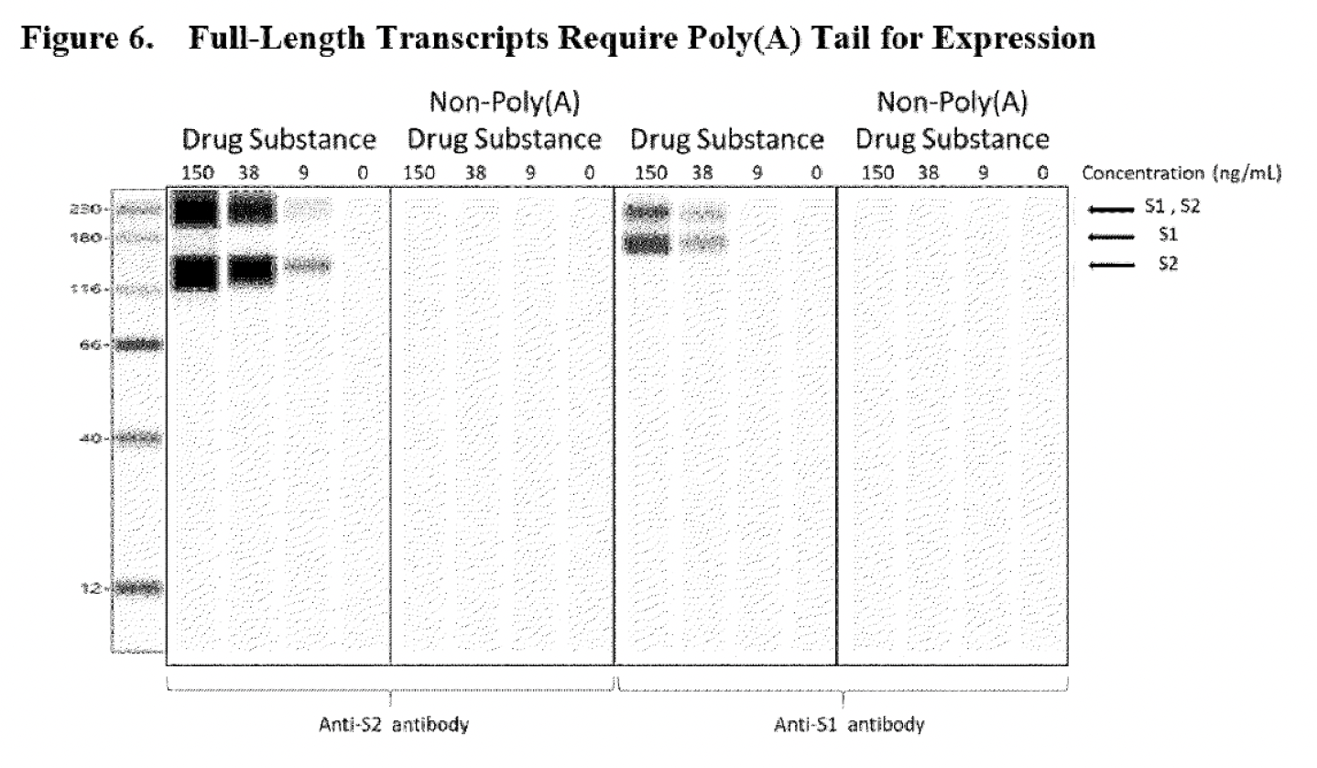
The blank lines shown in the non-poly(A) drug substance panels, were sufficient to prove to the regulatory authority that RNA lacking Poly(A) would not be able to express the S1S2 spike protein. However, the problem with this assay is that it does not confirm that any other aberrant proteins (which was an initial concern of the EMA) could be expressed by this truncated RNA because only S1 and S2 domain-specific detection antibodies were used. The Western blot shown below shares the same problem. This assay was used to reassure the regulatory authority that the RNA transcript lacking a 5’cap could not express the S1S2 spike protein.
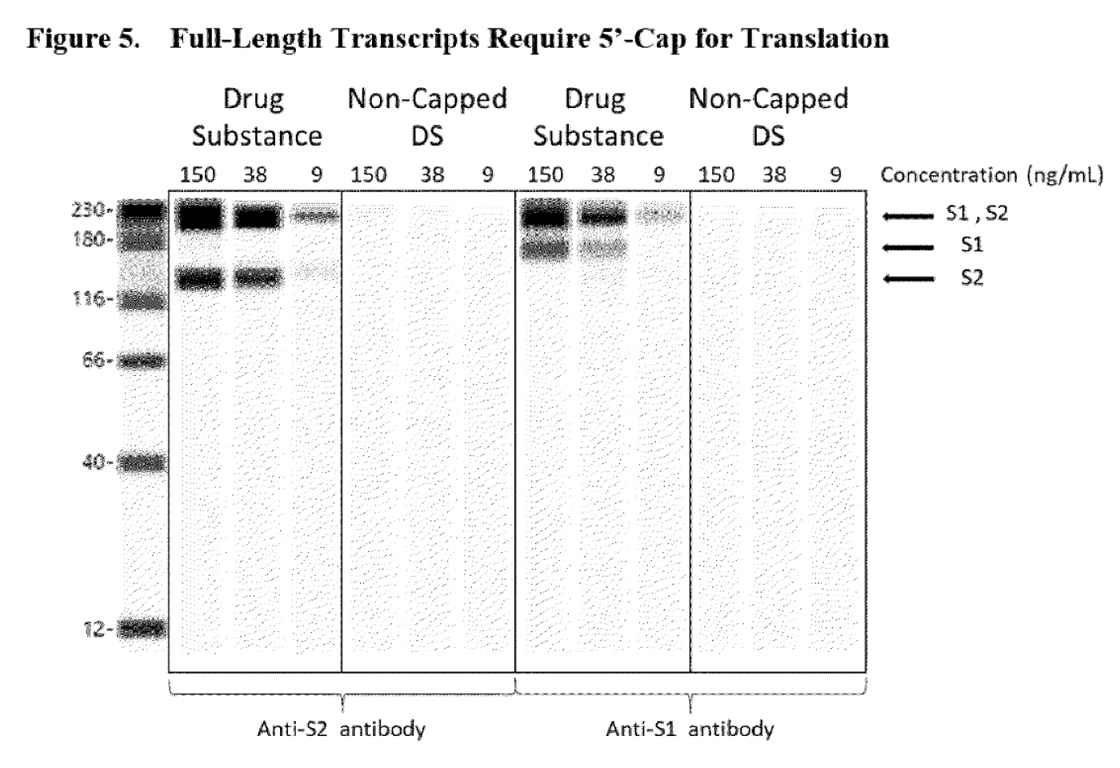
In addition, it was assumed that the fragmented RNA species was the result of premature transcriptional stops or mRNA hydrolysis (when a molecule breaks into two pieces when reacting with water), see excerpt below.
‘The associated safety assessment showed that the likelihood of fragmented species was generated by premature transcriptional stops or mRNA hydrolysis. As such, fragmented species predominantly lack both the 5’-cap and poly(A) tail elements required for protein expression
Source: EMA CHMP report of August 2021
This gave a false sense of security that a truncated mRNA transcript might only have either a 5’cap or a poly(A) tail, but never both.
The mirage of the degradation test
The potential of truncated RNA transcripts to produce proteins was further investigated by BioNTech at the request of the regulator. What is interesting is that the batch (1071509) was intentionally selected and intentionally degraded by exposure to high temperatures to generate samples with fragmented species. The Western blot shown below is the second traditional one submitted by BioNTech. It shows that the ability of the RNA transcript (when degraded by heat, hence fragmented) to express a protein becomes lower.
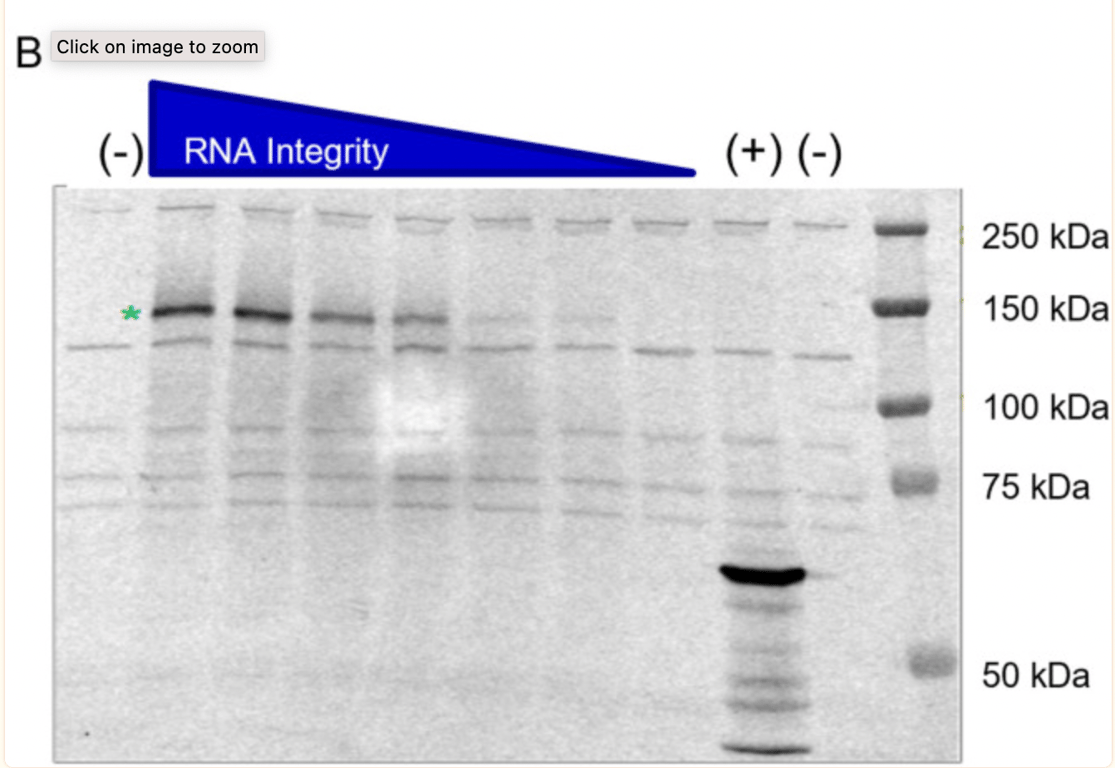
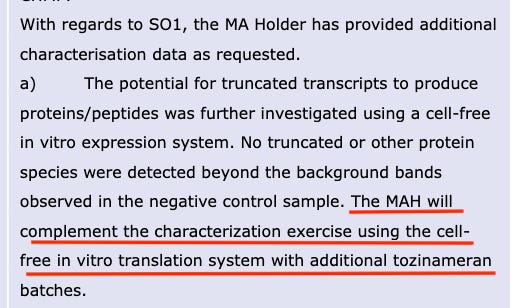
The green star shows that the non-heat degraded BNT162b2 sample (no heat, intact/high integrity RNA) produced a protein of about 140kDa, consistent with the expected size of the aglycosylated S1S2 protein. BioNTech states that “No truncated or other protein species were detected beyond the background bands seen in the negative control sample”, however, in the band of the undegraded sample, protein bands are visible and no explanation is given as to what they are.
In McKernon’s interview, he explained, “what they did in their study is they took mRNA and they didn’t fragment it like you get from the manufacturing process, they heated it! The reason they’re fragmented is that the polymerases get stuck on these non-native (modified) bases in RNA synthesis, so you get these shorter portions of RNA in the manufacturing process, and that process probably varies depending on the nucleotides they get from their suppliers”
In the August 2021 EMA report, a new requirement was made for BioNTech to fulfill- a request for the same characterization exercise to be done for at least three additional batches of tozinameran (modified mRNA, drug substance). As you can see, this has not been met by August 2021.
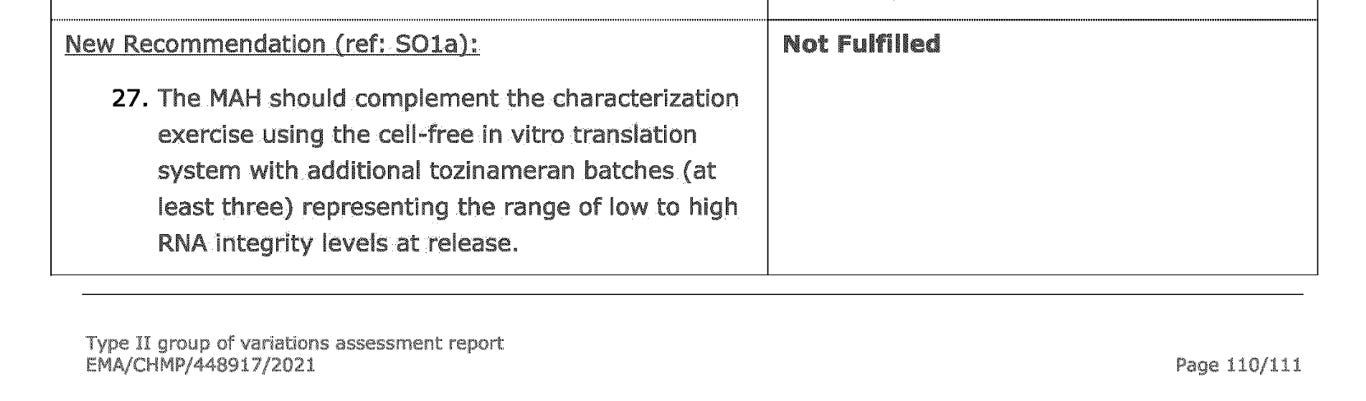
And according to the latest update (2 February 2023) Comirnaty: EPAR report, this obligation still remained unfulfilled.
McKernan also referred to the work of Patterson et al. which, alarmingly, found mutant versions of the spike protein in vaccinated people that did not exist in unvaccinated people who had COVID-19. This can be taken as real-world observational data showing that mutant (aberrant) versions of the spike protein are translated by vaccine-modified mRNA.
In his closing comment, McKernan said, ” Regulatory people are asleep at the wheel. They have left the helm and let it go on cruise control”
I couldn’t agree more.
Originally published on Trial Site News
Suggest a correction






